+66 2 5646700 Ext 71441 | noc.th@nstda.or.th

ศูนย์โอมิกส์แห่งชาติ

ศูนย์โอมิกส์แห่งชาติ
Bioinformatics
We have an excellent team of experienced bioinformaticians with expertise in de novo genome assembly, gene annotation, RNA-seq expression analysis, linkage map construction, marker-trait association analyses, metagenomic analysis and comparative genomic analysis. We have been working with organisms of various genome sizes from bacteria (~5 Mb), fungi (~40-100 Mb), animals (1-3 Gb) to plants (~200 Mb to 10Gb!)
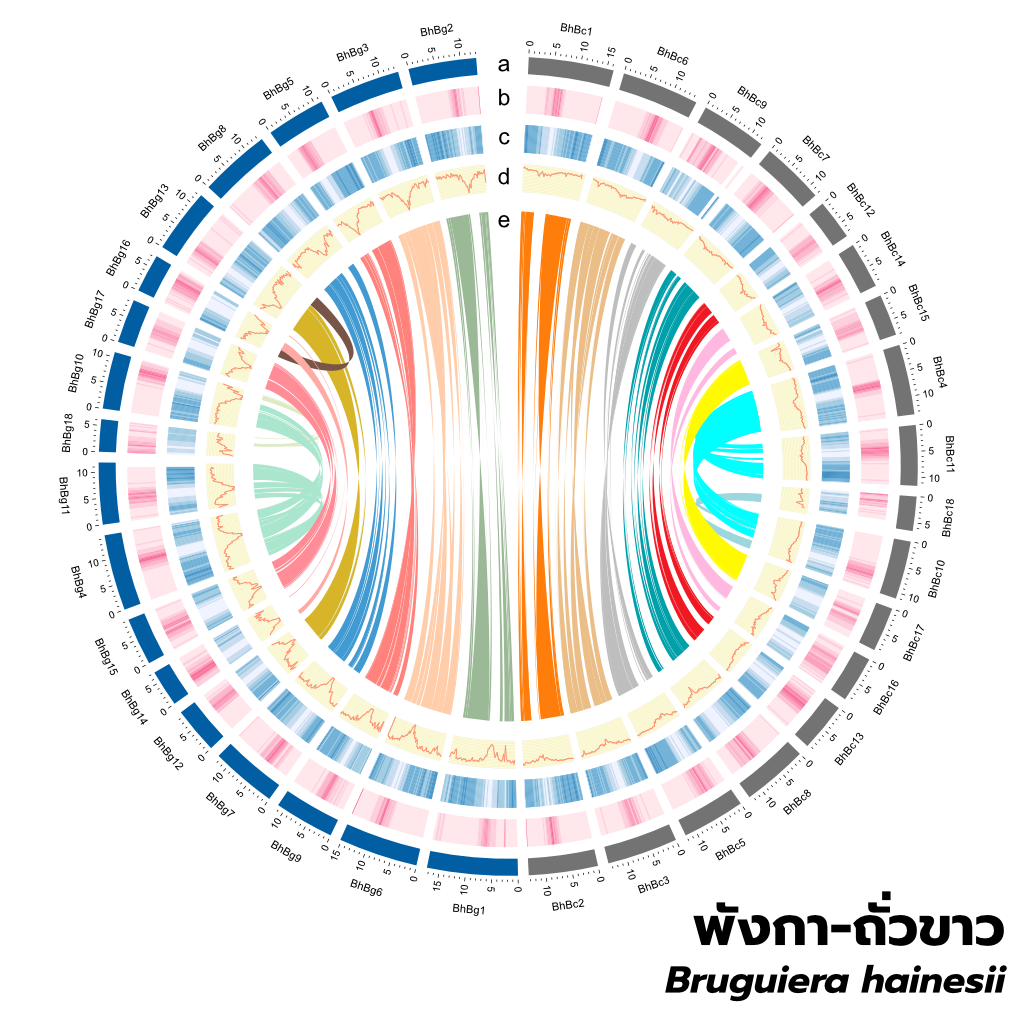
Genome Assembly: Our genome assembly strategy leverages the advantages of both short- and long-read sequencing. Short reads provide high accuracy, while long reads span repetitive regions, facilitating contig assembly. Subsequent Hi-C scaffolding then links these contigs into chromosome-scale scaffolds, producing near-complete genome assemblies. We have applied this technique to a wide range of plant and animal species, demonstrating its scalability from the compact chloroplast genome to the immense and complex genome of octoploid sugarcane.
Gene Annotation: To generate a comprehensive genome annotation, we employ a robust pipeline. This pipeline leverages RNA-seq data, ab initio gene prediction, and protein homology from related species to construct consensus gene models. Subsequent annotation is performed using Blast2GO. Repeat sequences are identified and annotated with RepeatModeler. Finally, all data layers are integrated into a GFF3 file, providing a fully annotated genome.









DNA fingerprint: Through SNP analysis, we identify a select set of highly informative SNPs capable of reliably differentiating between breeds, strains, or cultivars. These markers form the basis of a simple and inexpensive assay, enabling the screening of numerous samples for genetic origin and classification.
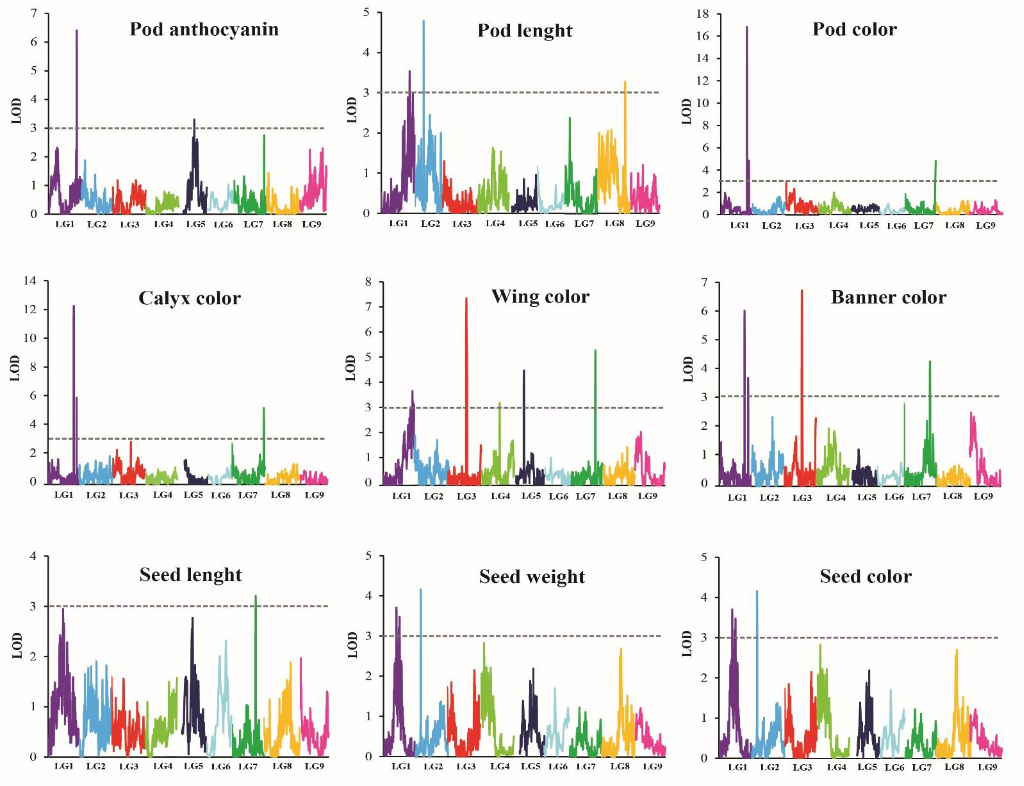
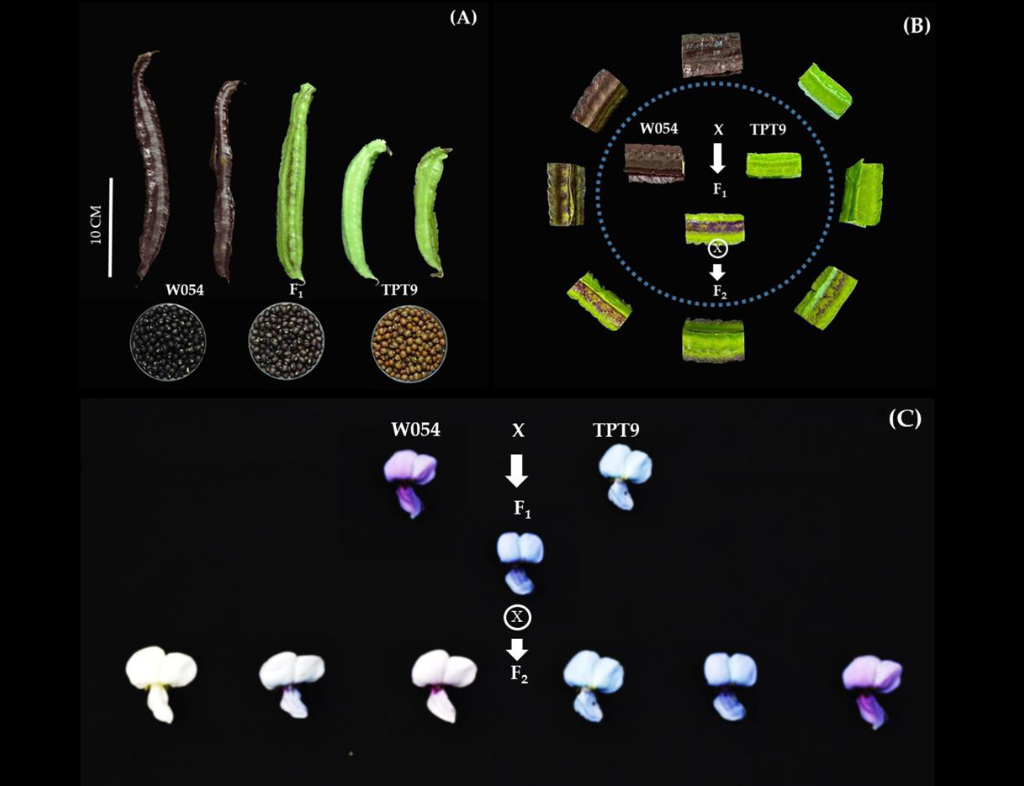
Trait – Marker Association Analysis: We leverage SNPs to identify genetic markers linked to traits of interest. Depending on the specific population structure, we employ either linkage analysis or quantitative trait locus (QTL) analysis to determine which markers occur with the trait more frequently than expected by chance. Once these regions are identified, we explore the genomic sequence to pinpoint candidate genes with functions relevant to the trait for further investigation.

Gene Expression Analysis: We use RNA-seq to profile the transcriptome of each sample. The proportion of reads mapping to a given gene provides a measure of its expression level. This allows us to compare gene expression levels between replicates of cases and controls (triplicate or higher recommended), or to track changes in expression over a time course.
SNP Identification: Two approaches are used for SNP identification: reduced-representation sequencing and whole-genome sequencing. Reduced-representation sequencing employs restriction digests to target a consistent subset of DNA fragments across samples, while whole-genome sequencing captures all genomic variation. In both methods, sequencing reads are mapped to a reference genome (or assembled de novo if necessary), and SNPs are called using GATK. The ability to process numerous samples allows for a wide range of analyses.
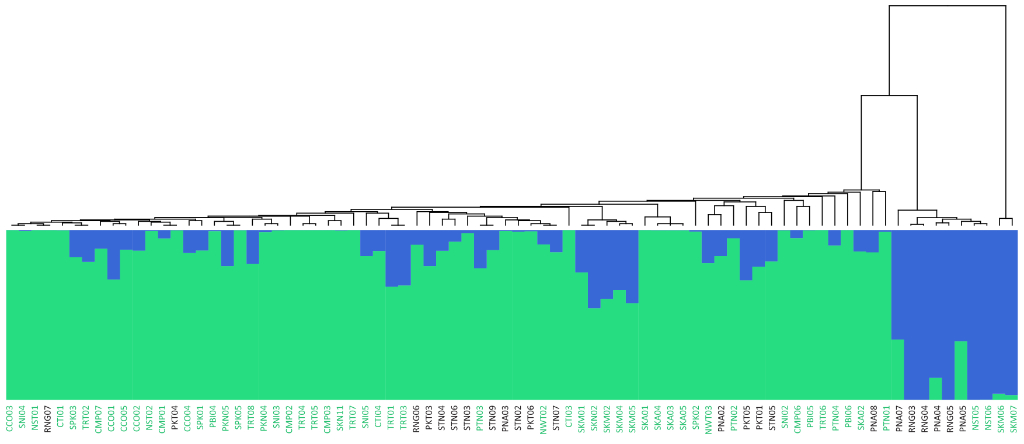
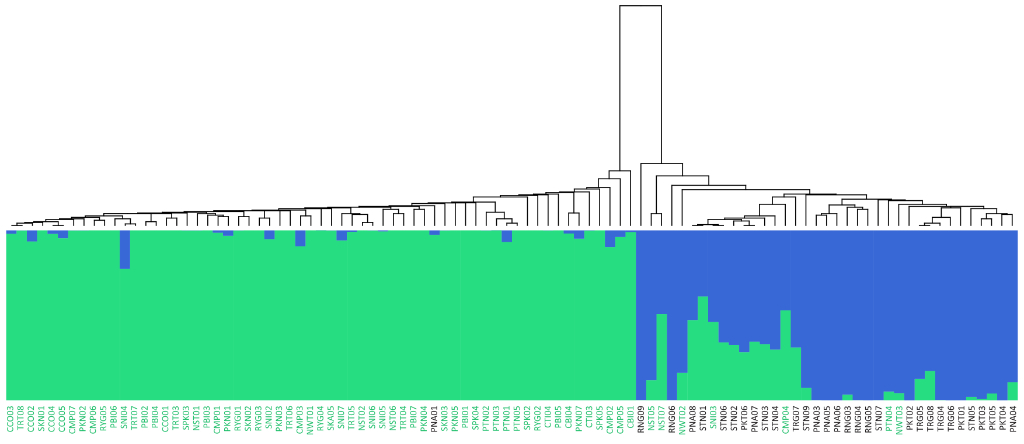
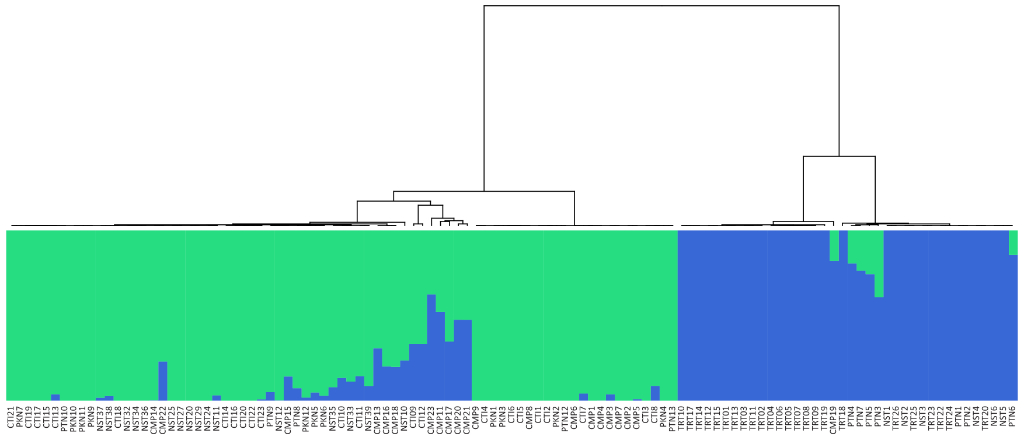
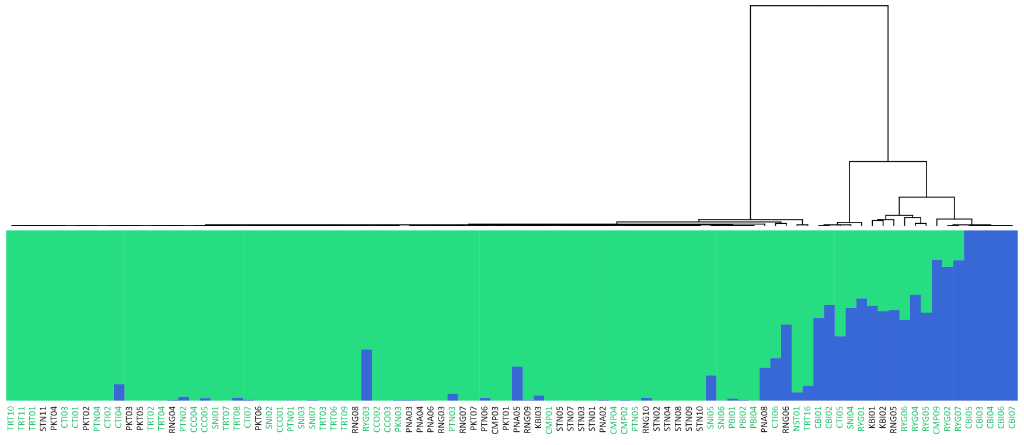
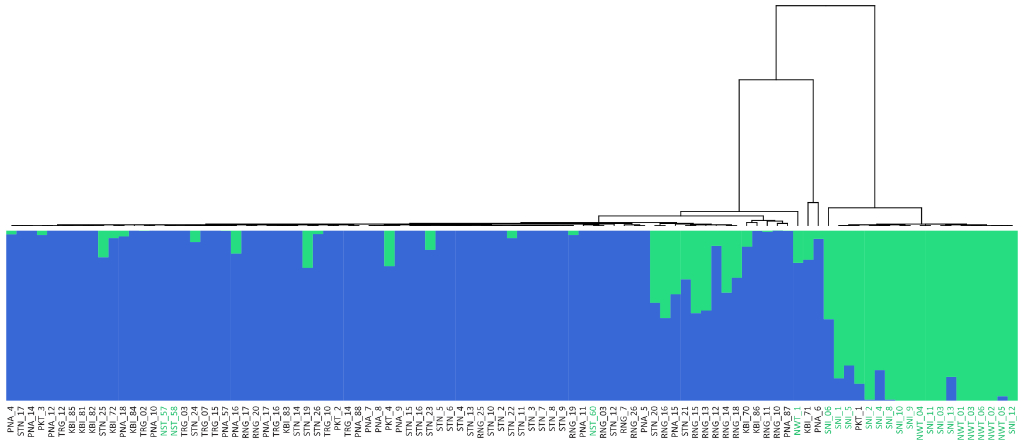
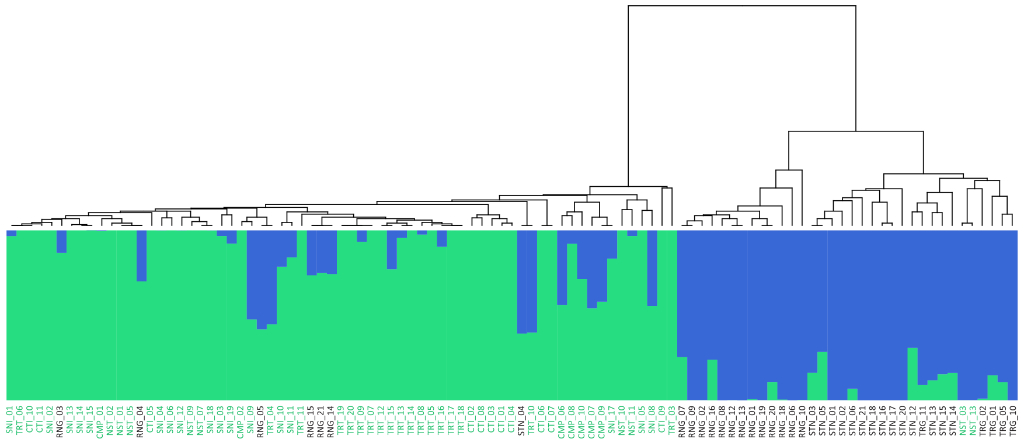

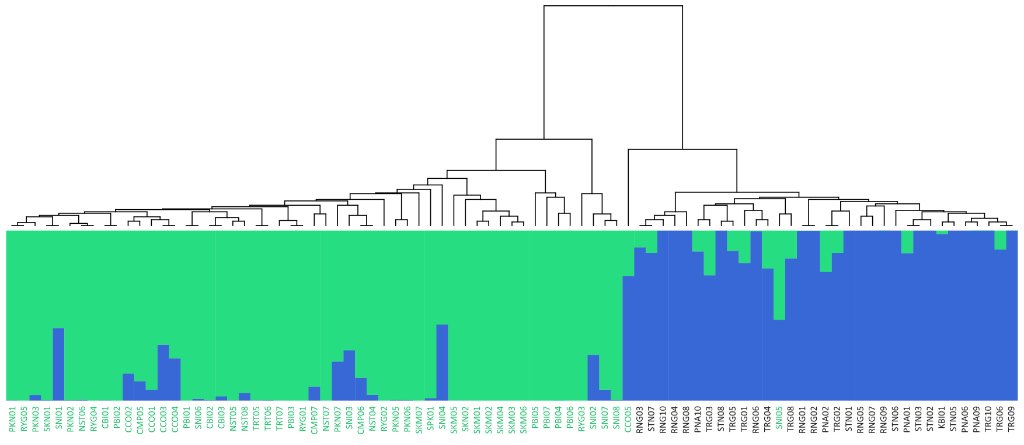

Population Genetic Analysis: Our population genetics service provides comprehensive analysis of your samples, revealing insights into genetic diversity, population structure, and evolutionary history. We offer customized solutions, from SNP genotyping and data analysis to the development of diagnostic markers and the interpretation of complex population dynamics. Whether you’re studying conservation, agriculture, or human populations, our expertise and cutting-edge tools deliver the data and insights you need to advance your research or breeding programs.
Metagenomics: We offer comprehensive metagenomics services, from sample preparation and sequencing to bioinformatic analysis and interpretation. Our expertise encompasses 16S rRNA gene sequencing for microbiome profiling, whole-metagenome shotgun sequencing for in-depth functional analysis, and custom bioinformatics pipelines tailored to your specific research questions. We deliver high-quality data and actionable insights, enabling you to explore the complex world of microbial communities and their impact on health, environment, and industry.
We offer comprehensive metagenomics services, from sample preparation and sequencing to bioinformatic analysis and interpretation. Our expertise encompasses amplicon sequencing with marker genes for microbiome profiling, shotgun metagenome sequencing for comprehensive functional analysis, and custom bioinformatics pipelines tailored to your specific research questions. We deliver high-quality data and actionable insights, enabling you to explore the complex world of microbial communities and their impact on health, the environment, and industry.
